Posts
Authors
- NIHR Maudsley BRC
- NIHR King's Clinical Research Facility
- NIHR King's Clinical Research Facility
- NIHR King's Clinical Research Facility
- NIHR King's Clinical Research Facility
- NIHR Wellcome King's Clinical Research Facility
Categories
Tags
- Publications (98)
- Informatics (69)
- Staff News (62)
- NIHR Wellcome King's Clinical Research Facility (57)
- Patient and Carer Involvement and Engagement (56)
- Clinical disorders and health behaviours (51)
- CRIS (47)
- South London and Maudsley NHS Foundation Trust (44)
- Covid-19 (43)
- Clinical and population informatics (32)
- Biomarkers & genomics (29)
- Eating Disorders & Obesity (26)
- Psychosis and Mood Disorders (26)
- Affective Disorders and Interface with Medicine (26)
- Pain and Addictions (25)
- research stories (23)
- Precision psychiatry (23)
- Events (22)
- Child & Neurodevelopmental disorders (22)
- Neuroimaging (21)
- BioResource (21)
- Dementia and related disorders (21)
- depression (21)
- Digital Therapies (20)
- Child Mental Health and Neurodevelopmental Disorders (18)
- Psychosis & neuropsychiatry (18)
- Substance use and harms (17)
- Training & capacity development (16)
- Public engagement (15)
- Equality Diversity and Inclusion (EDI) (14)
- Motor Neuron Disease (13)
- CRIS blog (12)
- BRC Interview Series (12)
- Novel Therapeutics (11)
- Experimental Medicine and Novel Therapeutics (9)
- Mobile health (9)
- King's Clinical Research Facility (8)
- Pain (8)
- Bioinformatics & statistics (8)
- Air Pollution (4)
- Geonomics and Prediction (3)
- Changing clinical practice stories (2)
- Community outreach (1)
- Award (1)
- equality diversity and inclusion (EDI) (1)
Archive
-
2025
- January
-
February
- New Deputy Director of NIHR Maudsley BRC announced
- Co-development of patient reported-outcome measures
- New MRI study reveals altered brain activity in depressed adolescents while watching ‘Despicable Me’
- APPG on MND Explores UK MND Research Institute’s Groundbreaking Work
- eLIXIR BiSL - How early life data is changing our understanding of health
-
March
- Genomic links between eating disorder symptoms and suicidal ideation
- Three quarters of people who have taken antidepressants say they were helpful
- EDGI UK collaborating with the Broad Institute to sequence exomes of participants with anorexia nervosa and bulimia nervosa
- Does the ethnicity of mental health research participants reflect the eligible population?
- Biological pathway in the brain could help explain why teenage girls are more depressed than boys
- April
-
May
- Groundbreaking AI trained on de-identified patient data to predict healthcare needs
- King's CRF Lead Nurse wins prestigious award
- New research finds Motor Neuron Disease drug safe and effective at low doses
- Ubrogepant found to be effective at reducing non-headache related symptoms of migraine
- Who benefits from antidepressants? Using medication switching as a measure of non-response
-
June
- King's CRF lead nurse awarded leadership scholarship
- Weight loss behaviours missing in tools to diagnose eating disorders
- Experts with lived experience address mental health stigma
- Bringing clinical research to the local community
- RE-STAR paper co-authored with Youth Researcher Panel wins British Journal of Psychiatry Editor's Choice Award
- Dani Nebres shortlisted for a Nursing Times Award 2025
- New paper showcases the work of the CAMHS Digital Lab as part of transforming child mental health care systems
- July
-
2024
-
January
- Professor Ulrike Schmidt recognised in New Year Honours
- Volume of grey brain matter significantly lower in people with Early Onset Psychosis
- An interview with... Professor Richard Emsley
- Deputy Director for NIHR King’s Clinical Research Facility (CRF) appointed
- Pioneering dance performance explores ‘living well with technology’ at King’s Chapel
- Mood interventions may reduce inflammation in Crohn’s and Colitis
- Clinician and patient views on a new rheumatology drug: Article published by a Patient Researcher
-
February
- Study estimates number of patients for potential new Alzheimer’s disease treatments
- Pioneering link between census data and electronic mental health records
- Patterns of brain connectivity differ between pre-term and term babies
- King’s College London announced winner of the Government’s AI Fairness Innovation Challenge
- FREED launches online training resources to ease transition from child to adult eating disorder services
- New digital therapy reduces anxiety and depression in people living with long-term physical health conditions
- King’s led Multiple Sclerosis fatigue app part of government’s £10M medtech investment
- March
- April
-
May
- Professor Gráinne McAlonan appointed as new Director of the NIHR Maudsley BRC
- Day workshop in Cognitive Behavioural Therapy effectively reduces depression in 16-18 year olds
- Why diversity in nature could be the key to mental wellbeing
- £4.8 million Wellcome funding to predict outcomes following anxiety treatment
- Self-harm and digital technology overuse in young people with lived mental health experience
- Ancient viral DNA in the human genome linked to major psychiatric disorders
- Depression, schizophrenia and bipolar disorder linked with ancient viral DNA in our genome – new research
- New Doctoral Training Centre to investigate Multiple Sclerosis (MS) symptom management
- Professor Ammar Al-Chalabi elected as Fellow of the Academy of Medical Sciences
- June
- July
- August
- September
-
October
- People with severe mental illness more than four times as likely to die from pneumonia compared to the general population
- “Don’t feel like you have to push through”
- First study to show high potency cannabis use leaves unique signature on DNA
- Ancient viral DNA in the human genome linked to multiple sclerosis and amyotrophic lateral sclerosis
- Multi-site trial uses digital avatars to effectively reduce distressing voices in psychosis
- How a framework for remotely enabled co-design benefits a wider project
- HIBISCUS study: Exploring the use of a new drug in patients with sickle cell disease
- November
-
December
- NIHR Maudsley BRC: A Year in Review 2024
- “It is my way of giving something back to an NHS that has served me so well”
- Cannabis use increases risk of psychosis independently from genetic predisposition
- CEDI Impact Award winners announced
- Dr Alfredo Iacoangeli receives the 2024 Paolo Gontijo Award
- Addressing the unmet mental health needs of patients with chronic conditions
- “Missing link” between brain and body inflammatory signals identified in the skull
- Living alone is linked to poor health and unemployment amongst those with severe mental illness, study finds
- Researchers compare artificial intelligence ‘ageing clocks’ to predict health and lifespan
- UK Disability History Month: Being neurodivergent and living with Tourette's
- UK Disability History Month: Living with reduced mobility, bipolar disorder and other disabilities
-
January
-
2023
-
January
- New framework aims to unlock the potential for social media research into young people's mental health
- Relatives' experiences of violence from people living with severe mental health conditions: a neglected and poorly understood issue
- UKCRF Network awarded £2.4 million in public funding to support the delivery of early phase research studies
- £1.3 million Wellcome funding awarded to SlowMo
- February
- March
-
April
- New app could help more patients access treatment for psychosis
- Breaking the barriers between offenders and academia
- Researchers uncover new biological mechanisms underlying the link between childhood trauma and psychosis
- 'Our Research Themes': introducing our ten themes and the people leading them
- Experts update global guidelines on treating eating disorders with medication
- Higher rates of HIV diagnoses found amongst mental health service users
- 60 seconds interview with Dani Nebres
-
May
- 60 seconds interview with Angelo Jornacion
- Interview with Emily Lam for International Nurses Day
- Molecular ageing clocks – making the links between mental illness and shorter lifespans
- Dementia Awareness Week: An interview with Miguel Vasconcelos Da Silva
- New CEDI Champion for the NIHR Maudsley BRC
- Professor Ulrike Schmidt elected as Fellow of the Academy of Medical Sciences
- New Deputy Director of NIHR Maudsley BRC announced
- £42m boost to mental health research announced by Office for Life Sciences
- More depressed patients than previously estimated could have increased activation of their immune system
-
June
- Working with students and teachers to evaluate secondary school stress workshops
- Celebrating research with the launch of a new Take Part in Research online database, and lighting up buildings for #Red4Research day
- Antidepressants prescription associated with a lower risk of testing positive for COVID-19
- UK funders invest £4.25 million into new eating disorders research
- July
- August
-
September
- Inflammation may influence weight loss surgery outcomes, new study reveals
- Critical care admissions for people with dementia are increasing for those in their last year of life
- From Patient to Advocate: A patient's journey of research participation and passion for Equality, Diversity and Inclusion
- King’s innovative AI projects funded to tackle fatal opioid overdoses
- Inflammatory signs for adolescent depression differ between boys and girls
- East and South East Asian Heritage Month: an interview with Michelle
- Lord O’Shaughnessy visits King’s College London and the NIHR King’s Clinical Research Facility
- World Alzheimer's Day: An interview with Miguel Vasconcelos Da Silva
-
October
- Major advance for detecting brain conditions in babies
- What can humans do to guarantee an ethical AI in healthcare? (Blog: Part I)
- What can humans do to guarantee an ethical AI in healthcare? (Blog: Part II)
- People with severe mental illness at 50 per cent higher risk of death following COVID-19 infection
- £14m provided for research into the brain development of children who were "in the womb" during the COVID-19 pandemic
- Children with ADHD frequently use healthcare service before diagnosis, study finds
-
November
- Empowering voices: A public member's story in the fight against Mouth Cancer
- International Stress Awareness Week: Insights from a public member
- Brain health in over 50s deteriorated more rapidly during the pandemic
- King's CRF Nurse Team Lead: Why I love research
- A showcase of research in eating and weight disorders celebrates the launch of CREW
- New Psychedelics and Mental Health Research Centre Officially Opens
- Interview with Professor Mitul Mehta, Director of the Centre for Innovative Therapeutics (C-FIT)
- Disability History Month (16 November - 16 December)
- Breaking down barriers: Advancing gender equity in psychiatry
- December
-
January
-
2022
-
January
- New study demonstrates link between brain chemical and visual processing in autism
- Psilocybin, in 10mg or 25mg doses, has no detrimental effects in healthy people
- No convincing scientific evidence that hangover cures work, according to new research
- Next generation of rapid-acting antidepressants: Can ketamine help prevent suicide?
- New collaboration with digital therapeutics company to investigate preventable opioid respiratory deaths
- Bringing together imaging and genetic mapping to investigate patterns of vulnerability in the brain
- Love Your Liver Month
- A virtual reality ‘Shopping Task’ could help test for cognitive decline in adults
- Genetics helps estimate the risk of disease – but how much does it really tell us?
-
February
- CUES-Ed: Promoting mental health and wellbeing among primary school children
- Involving more voices in research through the Race and Ethnicity Advisory Group
- My JournE: Smartphone-based routine mental health monitoring in schools
- MyHealthE: Digital Innovation in Child and Adolescent Mental Health
- Novavax COVID-19 jab trialled at King’s CRF approved by UK drugs agency
- Lessons learnt as addictions researchers during a global pandemic
- Young women with epilepsy at greater risk of stress induced seizures and drug resistance
-
March
- New research suggests a causal link between blood group and severe COVID-19
- Telling our stories of research through a new microsite
- New centre to accelerate psychedelic research and models of care for mental health in the UK
- VIEWER platform receives high commendation for Best Mental Health Partnership with the NHS at 2022 HSJ Partnership Awards
- New Equality, Diversity and Inclusion Champion appointed
- Topping Out ceremony marks a milestone for The Pears Maudsley Centre for Children and Young People’s Mental Health
- April
-
May
- NIHR Maudsley BRC researchers lead first study of psilocybin in adults with autism
- Loneliness – so obviously important but very under-reported
- Professor Peter Goadsby elected to the Fellowship of the Royal Society
- Professor Peter Goadsby elected to the Royal Society Fellowship
- My research journey: Jemma's story
- Making referrals to the AVATAR2 trial: An interview with Dr Helen Harding
- Unplanned hospital admissions for people with dementia higher in the last year of life
- #BePartOfResearch campaign: Our activity
- June
-
July
- We need to put equity at the heart of pain management
- People with depression “stagnating” in primary care, says top UK Psychiatrist
- Hidden figures: Unveiling differences in diabetes care in people living with mild and moderate-to-severe dementia
- Bringing experience and research together to understand psychosis
- New multi-centre clinical trial to investigate psilocybin therapy in anorexia nervosa
-
August
- BRC PhD students attend NIHR Academy Doctoral Research Training Camp
- South Asian Heritage Month - an interview with Pratik
- Mental health stigma on Twitter during COVID-19: service user perspectives
- In search of lost treatment: how mental health research can be part of patient care plans
- ‘Drug related deaths are avoidable’ - South London and Maudsley and King's College London mark International Overdose Awareness Day 2022
-
September
- Going with the flow: study shows canals help boost your mood
- The power of co-production: Building service user research skills with the Recovery College
- Professor Sally Marlow to be first Researcher in Residence at BBC Radio 3
- Diagnosis of a genetic cause in hundreds of people with motor neuron disease could be missed due to “arbitrary age limits and rules” on genetic testing, new study shows
- October
-
December
- Adults living in areas with high air pollution are more likely to have multiple long-term health conditions
- How to Do Research and How to Be a Researcher
- Healthcare workers in England experience PTSD at twice the rate of the general public
- Evaluating effectiveness of treatment for adults with chronic fatigue syndrome
- A Year in Review 2022
-
January
-
2021
-
January
- Fellows award for Dr Katherine Young for work on the mental health of young people during and after the pandemic
- NEWS2 evaluated for prediction of severe COVID-19 outcome in large international study
- SIREN study finds past COVID-19 infection provides some immunity for at least five months
- Interview with Miguel Vasconcelos Da Silva
- COVID-19 lockdown loneliness linked to more depressive symptoms in older adults
- The significant effect of lockdown on gambler’s mental health
- Antibiotic may improve outcomes for depression in people with low level inflammation
- Novavax trial reveals 89.3% effectiveness in preventing COVID-19
- Case studies
- February
-
March
- Deciphering the genetics behind eating disorders
- NIHR appoints Senior Investigators for 2021
- Depression and anxiety are associated with disagreement between patient and doctor assessments of psoriasis severity.
- Professor Peter Goadsby awarded world’s top Brain Prize 2021
- Interview with Dr Parisa Mansoori
- New study highlights the urgent need to reduce inflammation in overweight people with depression
- Novavax confirms vaccine provides 100% protection against severe COVID-19
- NIHR welcomes new vision for the Future of UK Clinical Research Delivery
- COVID-19 pandemic leads to rapid uptake of remote consultations in mental healthcare
- April
-
May
- An Interview with Chifundo Stubbs
- Brain scans could offer sign of postpartum psychosis risk
- Innovative UK data hub to enable research and innovation to tackle mental illness
- Inflammation is a core feature of depression: new evidence from large-scale study
- Mothers’ depression impacts mother-infant relationships
- Shifts to remote mental health services continued after lockdown, according to new study
-
June
- An interview with Zunera Khan
- Silent MRI: improving access to neuroimaging research
- Simple blood test can accurately reveal underlying neurodegeneration, according to new research
- New research from King's has identified three key inflammatory proteins which are lower in individuals at risk of severe COVID-19.
- New insight into how anti-inflammatory effects of omega-3 fatty acids could help reduce depression
- Five ways universities could improve mental health support for male students
- CogStack wins an Artificial Intelligence in Health and Care Award
- Volunteer calls for others to support dementia research in south London
- Europe's first 'game-changing' portable MRI machine arrives at King's Health Partners
- Global ALS/MND recognition Day
- Multiple long-term physical health problems increase risk of depression later in life
-
July
- An interview with Dr Marija-Magdalena Petrinovic
- COVID-19 variant vaccine begins recruiting in south London
- New Race and Ethnicity Advisory (READ) group is recruiting members
- NIHR Fellowship awarded to Dr Brendon Stubbs for research in persistent pain and serious mental illness
- Knowing what we don’t know: How statistics can help autistic people to live their best life
-
August
- New funding for innovative neuroimaging research
- Words don’t come easy: identifying perinatal self-harm in healthcare records
- Problems in thinking and attention linked to COVID-19 infection
- Opportunity for researchers to consult with new Race and Ethnicity Advisory group
- Exposure to air pollution linked with increased mental health service-use, new study finds
- Maudsley Hospital and King’s College London’s National Addiction Centre light up purple to raise awareness for International Overdose Awareness Day
-
September
- An interview with Charles Curtis
- Join our Adolescent Mental Health Advisory Group
- Common factors within the gut associated with depression and bipolar disorder
- SURE Recovery App – two years on after its launch
- Using CRIS to map the impact of the COVID-19 pandemic – service user and carer priorities
- Genetic risks for depression differ between East Asian and European groups
-
October
- Excess deaths in people with mental health conditions increased during the COVID-19 pandemic
- ADHD Awareness Month: Developing neurofeedback as a new treatment for ADHD
- Genetic risk of mental health conditions may influence where people choose to live, study suggests
- Eating disorders are just as likely to start in adulthood as childhood, report finds
-
November
- An interview with Dr Charlotte Tye
- NIHR: Your Path in Research
- Rett Syndrome Awareness Month
- Using MRI to investigate the brain response to inflammatory stimuli
- MANTRA training developed at SLaM to be rolled out to adult eating disorder services across the UK
- Six years of memory decline seen in anxious, depressed older people during pandemic
- An interview with Dr Katherine Young
- The silent and widely impacting cost of Vulvodynia: lessons learned and future recommendations
- What can research tell us about engaging adolescent boys into school-based mental health workshops?
-
December
- Diagnosis of depression in adolescents can negatively impact educational performance, study shows
- Using neurofeedback as a means of treating feelings of self-blame in depression
- Review looks at benefits and barriers following shift to remote mental health services during pandemic
- What is treatment-resistant depression? New report calls for clearer definition to inform research and improve treatment
- NIHR Maudsley BRC: A Year in Review 2021
- UK Disability History Month - an interview with Naomi
- People feel lonelier in crowded cities – but green spaces can help
- Vitamin D supplementation does not improve symptoms in people with psychosis, study finds
-
January
-
2020
-
January
- BRC researchers on the Highly Cited list doubles to twenty
- CRIS Blog: Are we under-estimating self-harm rates due to differences in hospital admittance procedures?
- BRC Researchers celebrated at the King’s Awards
- New study finds evidence for reduced brain connections in schizophrenia
- Exposure to trauma ‘activates’ genes into causing depression
- Nurses in research blog: Emma and Naomi
- New findings on the effects of cannabidiol on people with psychosis
-
February
- New centre of excellence for children and young people's mental health launched
- High volumes of mental health-related tweets associated with crisis referrals
- Call opens to drive the future of health data research
- CRIS Blog: Answering real-world questions about medication and mental health through pharmacoepidemiology
- CRIS Blog: Appropriate use of healthcare records for research
- CRIS Blog: Artificial Intelligence and Data in Suicide Prevention
- Largest ever study of eating disorders launches in England
- C4C research register now totals 20,000 people
-
March
- Georgia’s research secondment
- Study finds that Community Treatment Orders do not reduce hospital readmission rates or stays
- 10,000 people could benefit from new migraine drug
- Thirty risk factors found during and after pregnancy for children developing psychosis
- Heroin injection associated with respiratory disorder
-
April
- Researchers appeal to public for help to assess mental health impact of the COVID-19 pandemic
- CRIS Blog: CRIS in the time of coronavirus
- Depression and anxiety increase premature death by up to 134%
- Mental health and brain research must be a higher priority in global response to tackle COVID-19 pandemic
- Stories from our students: Becki
- COPE Study: Investigating the impact of the COVID-19 pandemic on mental health and well-being
-
May
- Thought provoking men’s mental health film released
- Animated parenting tips for struggling households
- Researchers track COVID-19 isolation effects on older people’s health and wellbeing
- From Babylonian blood-letting practices to wearable tech, new film on the evolution of depression perceptions and treatment
- Patient and public recommendations for getting involved in BRC research
- Cognitive behavioural therapy reduces the impact of dissociative seizures
- Mind the Gap 17-25: A diagnosis doesn’t define you
- Sarah Markham writes about her experiences in research
- Covid-19 Psychiatry and Neurological Genetics (COPING) study
-
June
- Trial testing a unique formulation of ibuprofen to treat COVID-19 launches
- New study to monitor the real-time effect of COVID-19 on mental health services
- Introducing the CRIS Natural Language Processing (NLP) Service
- Study shows Cognitive Remediation Therapy leads to improvement in cognitive skills and well-being in people with bipolar disorder
- New Research Training and Capacity Development lead announced
-
July
- Growing numbers of alcohol related hospital admissions linked to local spending cuts
- ACE inhibitors and ARBs not associated with severity of Coronavirus
- Data linkages animation explores the evolution of healthcare records in research
- Study estimates impact of COVID-19 pandemic on UK mental health after first month of lockdown
- Genes related to inflammation and stress may help tailor treatments for depression
- Can wearables like Fitbit devices be used to help detect COVID-19?
- Lithium in drinking water linked with lower suicide rates
- August
- September
-
October
- Loss of potential: teens diagnosed with depression show reduction in educational achievement from primary school to GCSE
- £1.2 million to roll-out dementia care home programme to COVID-hit sector
- NIHR announces mental health research goals for next decade
- Rosalind helps researchers navigate personal health data
- An interview with Dr Ndaba Mazibuko
- National study into neurological impact of COVID-19
- Study supports link between traffic-related air pollution and mental disorders
- How our eLIXIR research database helps reduce risk of health problems in mothers and children
- November
- December
-
January
-
2019
- January
-
February
- Eating a healthy diet can ease symptoms of depression
- CRIS Blog: Pathfinders and the public
- NIHR Maudsley BRC researchers host dementia discussion in collaboration with South London Theatre
- Could intranasal oxytocin be used to treat people at clinically high risk of psychosis?
- CRIS blog: Using data on hospital episodes to look at the physical health of people with personality disorders
- Cannabis-based medicine to be tested in Alzheimer's trial
- Largest ever study of depression and anxiety now recruiting individuals from Northern Ireland, Scotland, and Wales
- March
- April
- May
- June
-
July
- One in ten UK hospital inpatients is alcohol dependent
- CRIS Blog: Art and Value at Bethlem Gallery: an art-science collaboration with Sarah Carpenter and CRIS
- Genetic study reveals metabolic origins of anorexia
- Compensatory strategies to disguise autism spectrum disorder may delay diagnosis and have negative consequences for mental health
- Close monitoring essential to ensure safety of ketamine for depression
- August
- September
-
October
- CRIS Blog: CRIS data demonstrates need for better physical healthcare for people who use heroin
- SURE Recovery: the new addiction recovery app designed alongside service users
- Our first ever artist residency to kick off with Afrobeat and Dub gig in South London
- Expert panel examines barriers faced by working class academics
- December
-
2018
- January
- February
- March
- April
- May
- June
- July
- August
- September
- October
-
November
- CRIS blog: The future of psychiatry research
- CRIS blog: Do long-term prescriptions of multiple antipsychotics contribute to the reduced life expectancy of patients with serious mental illness?
- Improving dementia care and treatment saves thousands of pounds in care homes
- New service in south London reduces hospital readmissions for people with bipolar disorder
- Ten BRC researchers and academics among most cited in the world
- Students 'take over' the BRC
-
December
- Computers can ‘spot the difference’ between healthy brains and the brains of people with Dissociative Identity Disorder
- Service User Advisory Group for 12-16-year-olds
- New Clinical Disorders and Health Behaviours cluster lead announced
- Blog: The SLG Arts Assassins collaborate with the BRC
- Professor Matthew Hotopf receives CBE at Buckingham Palace
-
2017
- January
- February
-
March
- Professor Robert Stewart awarded ‘Collaborate to Innovate’ project
- NIHR Maudsley BRC researchers receive Senior Investigator awards
- Research blog: Using social media to recognise mental health conditions
- Department of Health Chief Scientific Adviser Chris Whitty visits Maudsley BRC
- NIHR Maudsley BRC commences five-year research programme
- IMPARTS Seminar Learning from experience
-
April
- New research highlights higher hospitalisation rates in people with intellectual disabilities
- Digital Technology for Mental Health: Asking the right questions
- Maudsley becomes London’s Global Digital Exemplar
- CRIS blog: An online risk calculator to identify candidates for early intervention services
- May
- June
- July
- August
-
November
- Professor Oliver Howes receives Royal College of Psychiatrists Award
- First network analysis of patient flow in two UK hospitals published
- Honorary Degree for Professor Dame Til Wykes
- Concentrated naloxone nasal spray as good as injection
- Potential for machine learning to predict unknown adverse drug reactions
- Complications at birth associated with lasting chemical changes in the brain
- Study examines opiate-dependent patient deaths
- NIHR Lectureship awarded
- Treatment cuts migraine days by half
- December
-
2016
- January
- February
- March
-
April
- CRIS blog: Eight years on
- Experts call for greater recognition of little-known forms of dementia
- Event: Clinical Research Facility Research Forum
- Al Chalabi Sheila Essey Award
- RADAR CNS smartphone wearable devices transform medical care
- Cardiovascular drug underprescribing
- Consultation reveals better integration between physical & mental health physicians as top priority
- Prestigious Fellowships awarded to BRC researchers
-
May
- BMJ Award for team who are incorporating mental health service into dermatology clinic
- Research blog systematic biases in death certification
- Victoria Derbyshire show inheritance mental illness
- BRC spin out Mindwave launches
- Research blog: Learning how to be a critical friend to researchers
- Research blog International Clinical Trials Day
- June
- July
- August
- September
- October
-
November
- South London and Maudsley rated top mental health trust for recruiting patients to clinical studies
- NIHR i4i mental health challenge launch
- UK Government announces £4m investment in NIHR Wellcome Trust King’s Clinical Research Facility
- NIHR Maudsley BRC PhD student wins International Society for the History of Neurosciences book prize
- NIHR Maudsley BRC's takeover challenge
- December
- 2015
Bringing together imaging and genetic mapping to investigate patterns of vulnerability in the brain
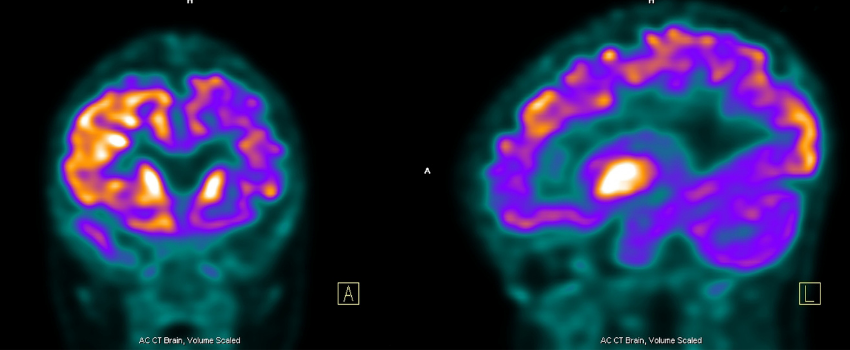
Dr Daniel Martins is a Clinical Research Associate, Dr Mattia Veronese is a senior molecular imaging scientist and Dr. Ottavia Dipasquale is a Research Associate, all in the Department of Neuroimaging at King’s College London. In this blog they discuss their recent study which is funded by the NIHR Maudsley BRC and has brought together scientists from across the world to define a new approach to integrate neuroimaging and a form of genetic mapping.
The emergence of non-invasive neuroimaging techniques, such as magnetic resonance imaging (MRI), and their increasingly large role in human brain research over the last decades have revolutionised the way we understand the brain and its dysfunction in disease.
Thanks to MRI and other imaging techniques, we can now describe a range of changes in the structure, function and chemistry of the brain in a variety of disorders, simply by acquiring images of patients from a scanner. In most cases, these data have made significant contributions to improve our understanding of the role of the nervous system in the patterns of behaviour and thinking shown in different mental health conditions.
Over the years we have come to appreciate that, for most cases, changes and differences in the brains of patients with different conditions are not random but follow certain spatial patterns where some regions of the brain are more severely affected than others. We call this regional vulnerability and if we can find out more about what factors contribute to these patterns then this could provide insight into developing and evaluating new treatments.
“A dance for two”: Bringing neuroimaging and transcriptomics together
While often thought of as a single organ, the brain encompasses a vast number of different regions which vary in their composition at both a cellular and molecular level. We are interested in whether regional vulnerability to a certain brain disorder is reflected in the landscape of genes and cells that make up a certain brain region. A new technique called Imaging Transcriptomics that integrates imaging with a form of genetic analysis might hold promise in investigating this question.
Transcriptomics is the analysis of the transcriptome which is collection of all ribonucleic acid (RNA) that is present in a cell, tissue or organ at a given time. RNA performs many functions within the cell, and studying the transcriptome provides insights into how genes are working.
While neuroimaging data offer a unique opportunity to map brain changes in patients at the macroscopic level, most techniques do not allow to specifically quantify genes or cells because they lack the spatial resolution. However, there has been a huge effort from the molecular biology research community to map the processes by which genes get turned on to produce RNA and proteins across multiple regions in post-mortem brains.
A good example of this is the work of the Allen Institute to create the Allen Human Brain Atlas (AHBA). The incredible anatomical detail of this atlas alongside the effort to image all these post-mortem brains with MRI have for the first time opened the possibility to map the fundamental activity of genes in the brain to MRI scans.
This impressive resource coupled with work from research groups, including our own at the NIHR Maudsley Biomedical Research Centre (BRC), has led to Imaging Transcriptomics becoming a popular method in neuroimaging studies. In simple terms, this approach tries to predict the regional variation of certain neuroimaging markers, such as changes in brain structure or function during disease, based on data on genetic expression in the post-mortem human brain as available in the AHBA. However, despite its increasing application, there are still some open questions about whether this framework could capture something biologically meaningful or is it just an interesting new approach?
This is the question that our recent paper in Cell Reports has attempted to answer. By using data from positron emission tomography (PET) imaging – a technique that uses radioactive compounds to trace molecules in the brain - we have tested how well the gene and cellular composition of the brain might be reflected in neuroimaging measurements at the macroscopic level.
PET provides a great opportunity to understand the cellular and genetic make-up of the brain since we know which molecules the tracer binds to and what type of cell populations it might target. If we assume the imaging transcriptomics approach is valid then we can formulate more precise hypotheses on the genes that should align with the regional distribution of a neuroimaging marker and test this.
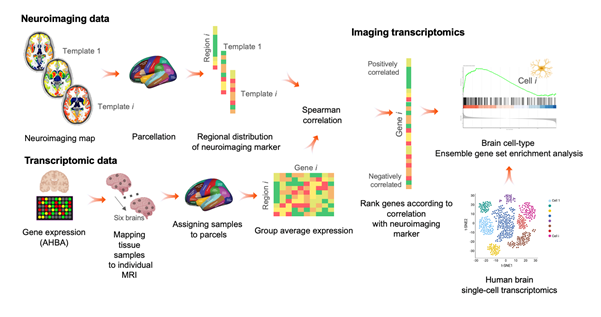 Imaging Transcriptomics framework (reproduced from Martins et al., 2021)
Imaging Transcriptomics framework (reproduced from Martins et al., 2021)
This was exactly what we did in our paper. By using several PET tracers, we could span a wide-range of different elements of the biology of the brain, from brain cells to star-shaped cells called astrocytes in the nervous system and myelin which is the layer around the nerve fibres.
With the exception of tracers that are currently used to study inflammation in the brain, we found that the expression of genes known to be present in certain types of brain cell could provide an efficient way to predict the extent to which a certain tracer binds in different regions of the healthy human brain. The reasons underlying the lack of correspondence between PET and genes that play a role in the inflammatory mechanisms in the brain is currently unknown, but we believe that the fact that we relied on PET data from healthy controls, where neuroinflammation is admittedly low, could have contributed.
The power of international collaboration and data sharing in medical research
One of the goals we had for this work was to investigate different molecular elements of the architecture of the human brain spanning different biological processes and cell types. To accomplish this mission, we needed data from a vast number of different PET tracers. This was beyond what our own library of PET templates included so to overcome this limitation, we contacted several research groups across the globe and setup a new initiative to gather PET data from healthy controls for 13 tracers through a new consortium – the PET templates Working Group.
Through this work, we created a unique resource which was only possible thanks to data sharing. We believe the resource will benefit further research in human neuroscience and has been a great example of the power and importance of collaboration in scientific research.
The future ahead…
Now that we are more confident that what we measure with neuroimaging reflects to an extent the gene and cellular landscape of the brain, there are several opportunities ahead.
For instance, by identifying genes whose expression preferentially align with the changes we see in neuroimaging data from a certain population of patients, we can now better inform hypotheses about genes that might contribute to the risk to disease and biological pathways that might contribute to the brain pathology. Ultimately, we hope that this approach can help identify certain molecular targets that might be worth exploring in studies for new therapies.
Tags: Biomarkers & genomics - Neuroimaging -
By NIHR Maudsley BRC at 21 Jan 2022, 09:38 AM
Back to Blog List




